Protein structures at fast MAS
Rapid data acquisition for de novo structure determination of proteins
We combine well-known techniques such as SIM-CP, co-evolution, bidirectional cross-polarization, orphan pathways and multiplexing, pioneered by several other groups in the field, to design proton-detected residue linking experiment. This sequence allows simultaneous acquisition of one new, CA(CON)CAHAand five previously known experiments NCAHA, N(CO)CAHA, CANH, CA(CO)NH and N(CACO)NH.48For the microcrystalline ubiquitin sample, all these spectra could be recorded in about 1.7 days compared to about 5-7 days of measurement time required to independently acquire all the spectra. These six spectra allow independent forward and backward backbone walks using the resolved 2D NHNand CAHAplanes. Rapid data acquisition combined with automated assignment makes this a compelling approach for de novo structural characterization of protein using nanomolar sample amounts.
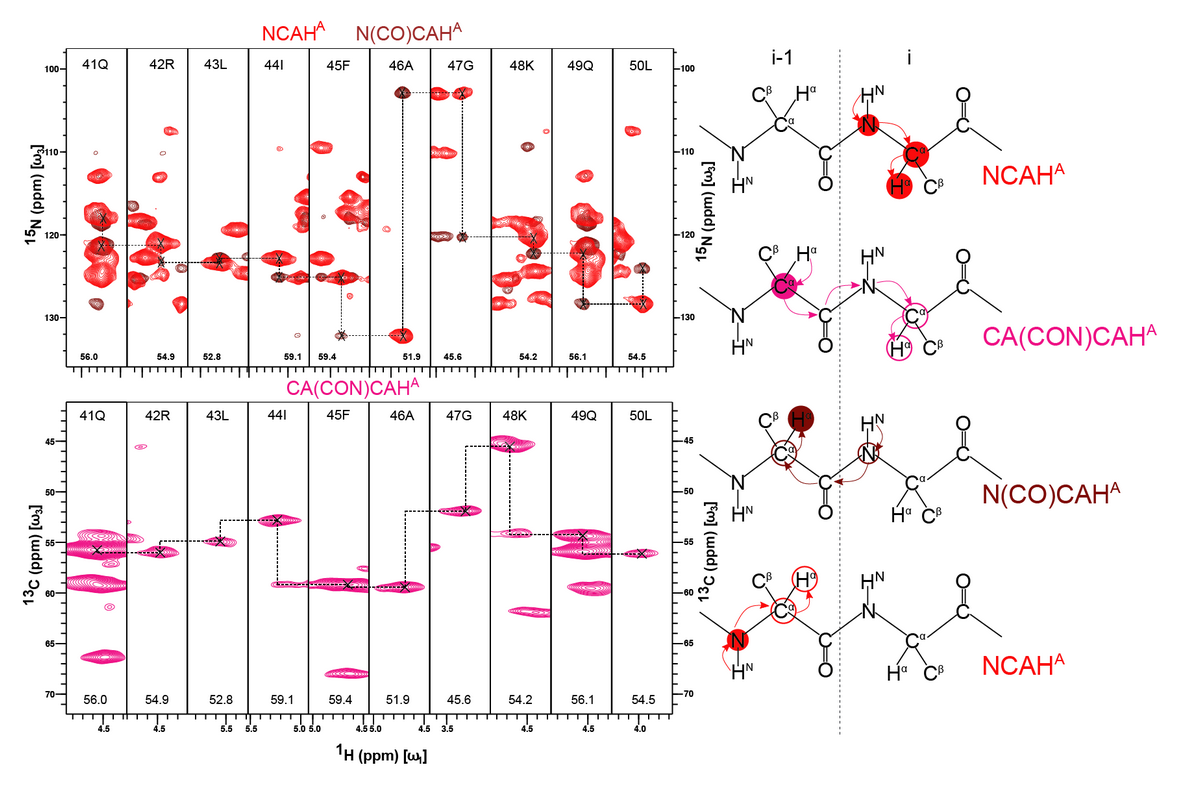
Figure: Two Independent backbone walks using two-dimensional HN and HaCa planes
This project is a collaborative effort with a colleague, Dr. Kaustubh Mote, at TIFR Hyderabad.
