Research-old
Our research focuses on understanding biophysically and biochemically interesting systems using novel tools of computer simulations and developing chemically motivated and physics-based model. We stress on using enhanced sampling methods to understand free-energetics and kinetics of diverse biologically and biochemically challenging systems. We also appreciate suitable collaborations from interested experimental groups.
The following research topics are of current interest :
1. Biophysics of protein-drug binding:
The efficacy of a drug depends on the efficiency at which the drug binds to the target protein. In many cases, the binding mechanism of drug gets perturbed by the mutations near the binding sites ( which is part of bacterial resistance) . Similarly, biological waters also play an important role in the binding process of the drug to the host protein. Understanding the processes of drug binding are challenging due to stochasticity and specificities involved in these binding events. We plan to apply novel computer simulation techniques to understand both thermodynamics and kinetics of a number of protein-drug binding (and unbinding) cases.
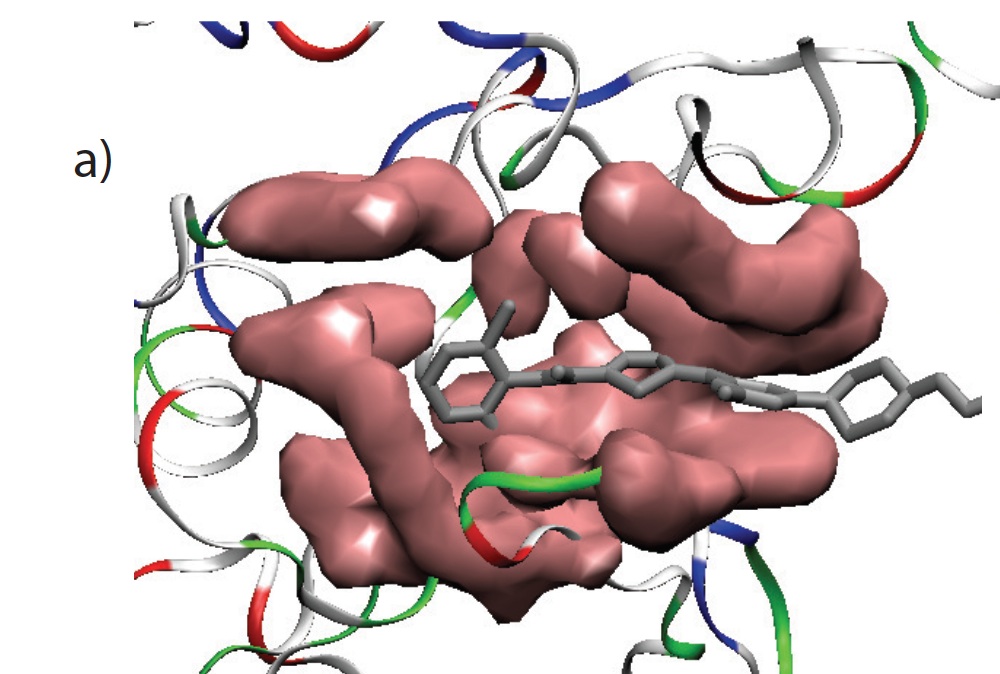
2. Understanding mechanisms of actions antibiotic and antimicrobial peptides:
Antibiotics and antimicrobial agents are two different avenues of disrupting bacterial host-defense systems. The mechanistic actions of these agents are complex and a full picture is yet to unwind. We aim to apply novel sampling techniques of computer simulations to decipher the mechanisms of action of antibiotic and antimicrobial peptides at an atomistic resolutions. We will focus on a popular antibiotic and a series of antimicrobial peptides. For antimicrobial peptides we will investigate the first step i.e. interaction of the antimicrobial agents with membrane. For antibiotics, we will investigate the role of amino-acid residues of the target protein in the interaction of protein-antibiotic actions.

3. Understanding role of water and cosolutes on conformation and self-assembly of biomacromolecules :
Under the realm of this topic, we will explore, using novel free energy simulation techniques, the conformational equilibrium of polymers and proteins. One of the thrust will be to identify, using tools of enhanced computer simulations, interesting collective variables to delineate the free-energy landscape of conformational equilibrium of the biopolymers and then perform kinetic-studies on these free energy surface. The study will be done in parallel with collaboration with some experimental group performing single molecule force spectroscopy. We will investigate the role of water, small cosolute molecules and the external biasing forces in perturbing the free-energetics and kinetics of the biomacromolecules.

4. Deciphering spatial organization inside bacterial cell:
The structure and dynamics of the bacterial chromosome are not well understood. A rapidly growing E. coli cell is a rod with length ~ 4 micrometer and diameter ~1 micrometer. The cytoplasm contains multiple genome equivalents of chromosomal DNA that occupy an irregular sub-region called the nucleoids. In spite of the large amount of DNA, the nucleoids do not fill the entire volume of the cytoplasm. The shape of the nucleoids may be governed by the ring topology of the nucleoids and spatial confinement effects on the ring polymer. The overall size of the nucleoids has been suggested to arise from a balance of nucleoid-expanding and nucleoid-compacting forces.

Despite its lack of organelles, the bacterial cytoplasm exhibits a high degree of spatial organization. The chromosomal DNA is condensed into a central region called the nucleoid, and specific DNA loci adopt specific locations during the cell cycle. In rapidly growing Escherichia coli and Bacillus subtilis, the ribosomes strongly concentrate in the cytoplasmic periphery, including the two poles (i.e., outside the nucleoid). Many proteins and even some lipids are known to have specific addresses in the cell. In this project, we plan to develop a multi scale model and use computer simulation to decipher the root of surprising self-organization of different biomacromolecules inside cell. Towards this end, we plan to build on our own previous polymer-physics based model to represent a Ecoli cell and some of its constituents. The questions that we would like to address will be the role of drugs on the organization of DNA and ribosome inside a bacterial cell, role of so-called Coupled transcription, translation, and insertion of membrane proteins (‘transertion’) mechanism and co-transcriptional translation mechanism in the DNA-ribosome organization and many other interesting biologically relevant questions.
